Conclusion 结论
Phage diversity operates differently depending on what aspect of phage biology is investigated. Nucleotide and gene content are extremely diverse in phage genomes, while protein structures are highly conserved among different phage families. We also highlighted recent work on viral metagenomics and how it contributed to the discovery of perhaps the most abundant phages in marine and gut environments. Viral metagenomics also expanded our knowledge of phage diversity in diverse ecosystems and the confines of this global diversity132. This ever-expanding catalogue of new phage sequences called for a reflection on how to best adapt the current viral taxonomy to properly classify phages discovered through metagenomics133. Phages are also interconnected from an evolutionary perspective and several factors drive higher or lower rates of gene exchanges. These complex phylogenetic relationships are more accurately represented by a network rather than a traditional tree, and the former may be better suited to define new phage genera, subfamilies and families16.
根据噬菌体生物学研究的不同方面,噬菌体多样性的作用也不同。噬菌体基因组中的核苷酸和基因内容极其多样,而不同噬菌体家族之间的蛋白质结构则高度保守。我们还重点介绍了病毒元基因组学的最新研究成果,以及它如何帮助发现海洋和肠道环境中可能最丰富的噬菌体。病毒元基因组学还扩展了我们对不同生态系统中噬菌体多样性以及这种全球多样性局限性的认识132。不断扩大的新噬菌体序列目录要求我们思考如何更好地调整当前的病毒分类法,以便对通过元基因组学发现的噬菌体进行正确分类133。从进化的角度来看,噬菌体之间也是相互关联的,有多种因素推动着基因交换率的高低。这些复杂的系统发育关系用网络而非传统的树形结构来表示更为准确,前者可能更适合定义新的噬菌体属、亚科和科16。
We also want to emphasize the success of the SEA-PHAGE educational program, which contributed to the isolation, characterization and sequencing of the largest collection of phages infecting the same host. In an era when a plethora of new phages with completely new sequences are being discovered, this model highlights the importance of integrating phage research at the various teaching levels, which benefits both the students and the scientific community. As more phages are discovered, the better the community will be at catching more of them and elucidating the viral dark matter. Adding more phage sequences to reference databases will help identifying a larger diversity of viral sequences from metagenomes. Resolving the structure of more viral proteins will also provide additional insights into the existence of a common ancestor, as intermediate ancestors within the lineage may be uncovered. Finally, network-based phylogenies will be improved when more phage sequences will be added, as it will help clustering more accurately phage groups that are poorly-sampled at the moment130.
我们还想强调的是,SEA-PHAGE 教育计划取得了成功,它促成了感染同一宿主的最大规模噬菌体的分离、定性和测序。在大量具有全新序列的新噬菌体不断被发现的时代,这种模式凸显了将噬菌体研究融入不同教学层次的重要性,这对学生和科学界都有好处。发现的噬菌体越多,科学界就越能捕捉到更多的噬菌体,阐明病毒暗物质。将更多的噬菌体序列添加到参考数据库中,将有助于从元基因组中鉴定出更多样的病毒序列。解决更多病毒蛋白质的结构问题也将为了解共同祖先的存在提供更多的信息,因为可能会发现病毒系中的中间祖先。最后,当更多的噬菌体序列加入时,基于网络的系统发生学将得到改善,因为这将有助于更准确地聚类目前采样不足的噬菌体群130。
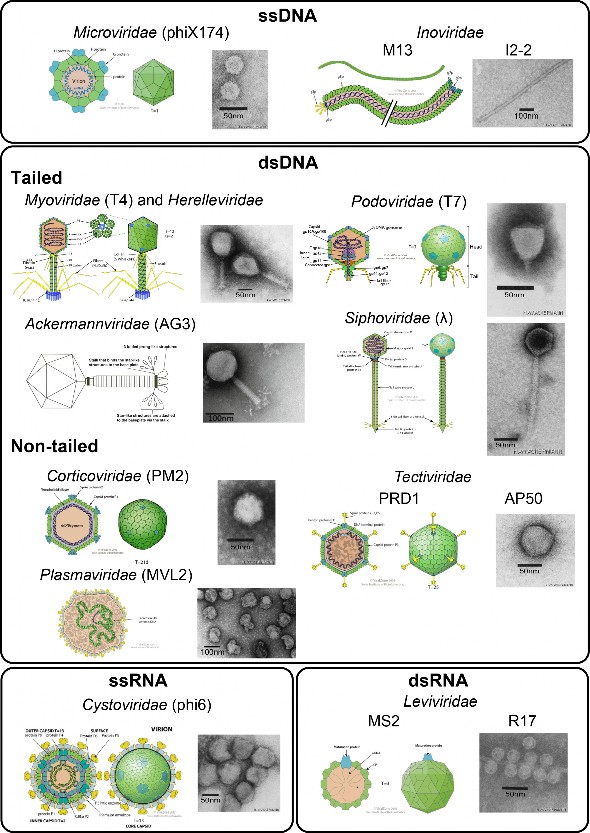
Fig. 1 Phage classification based on morphology and genome nucleotide type composition. A schematic representation (SR) and an transmission electron micrograph (TEM) are shown for each morphology. Microviridae have an icosahedral capsid and small circular ssDNA genomes (SR and TEM of phiX174). The genome of filamentous phages of the Inoviridae family is composed of a circular supercoiled ssDNA molecule which is packed in a long filament (>500 nm) composed of thousands of MCP13,134 (SR of M13 and TEM of I2-2). Most of the characterized phages are tailed with double-stranded DNA (dsDNA) genomes and belong to the Caudovirales order. To date, five families have been described for this order: Myoviridae (long contractile tails, SR and TEM of T4), Podoviridae (short non-contractile tails, SR and TEM of T7), Ackermannviridae (Myoviridae morphology with tail spikes at the base of the tail, SR and TEM of AG3) and Siphoviridae (long non-contractile tails, SR and EM of lambda). Herelleviridae, although an official family, shares the same morphology as Myoviridae and the two were merged in the figure. Corticoviridae have double-stranded circular DNA genome and capsids composed of an internal lipidic membrane surrounded by MCP (SR and TEM of PM2). Tectiviridae (SR of PRD1 and TEM of AP50) have an icosahedral capsid, which contains a linear double-stranded DNA genome and an internal lipidic membrane. Viruses belonging to the Plasmaviridae family (SR and TEM of MVL2) have a circular double-stranded DNA genome surrounded by lipidic envelope and no capsid. Cystoviridae have tri-segmented double-stranded RNA genomes contained in a spherical capsid (SR and TEM of phi6) with three structural layers: an outer lipidic membrane and a two layers inner capsid. Leviviridae have dsRNA genomes encoding only four proteins (MCP, replicase, maturation and lysis proteins) and capsids with icosahedral and spherical geometries (SR of MS2 and TEM of R17). Photo credit for EM goes to the late Prof. Dr. Hans-Wolfgang Ackermann (available at www.phage.ulaval.ca), with the exception of Ackermannviridae (from Adriaenssens et al. (2012) with permission) and Plasmaviridae (from Poddar et al. (1985) with permission).
图 1 基于形态和基因组核苷酸类型组成的噬菌体分类。图中显示了每种形态的示意图(SR)和透射电子显微照片(TEM)。微病毒科具有二十面体噬菌体和小的环状 ssDNA 基因组(phiX174 的 SR 和 TEM)。因诺病毒科丝状噬菌体的基因组由环状超卷曲 ssDNA 分子组成,该分子被包装在由数千个 MCP13,134 组成的长丝(>500 nm)中(M13 的 SR 和 I2-2 的 TEM)。大多数具有特征的噬菌体都带有双链 DNA(dsDNA)基因组,属于噬菌体目(Caudovirales order)。迄今为止,该目已描述了五个科:Myoviridae(长收缩尾,T4 的 SR 和 TEM)、Podoviridae(短非收缩尾,T7 的 SR 和 TEM)、Ackermannviridae(Myoviridae 形态,尾基部有尾钉,AG3 的 SR 和 TEM)和 Siphoviridae(长非收缩尾,λ 的 SR 和 EM)。Herelleviridae 虽然是一个正式的科,但其形态与 Myoviridae 相同,因此在图中将两者合并。皮层病毒科(Corticoviridae)有双链环状 DNA 基因组和由 MCP 包围的内部脂质膜组成的囊体(PM2 的 SR 和 TEM)。花叶病毒科(PRD1 的 SR 和 AP50 的 TEM)具有二十面体荚膜,其中包含线性双链 DNA 基因组和内部脂质膜。属于浆膜病毒科的病毒(MVL2 的 SR 和 TEM)具有环状双链 DNA 基因组,周围有脂质包膜,没有纤突。囊病毒科(Cystoviridae)的三节段双链 RNA 基因组包含在球形囊膜中(phi6 的 SR 和 TEM),囊膜有三层结构:外层脂质膜和两层内囊膜。Leviviridae 的 dsRNA 基因组只编码四种蛋白(MCP、复制酶、成熟蛋白和裂解蛋白),纤毛体呈二十面体和球形几何结构(MS2 的 SR 和 R17 的 TEM)。EM 的图片归功于已故的 Hans-Wolfgang Ackermann 教授(可在 www.phage.ulaval.ca 上查阅),但 Ackermannviridae(来自 Adriaenssens 等人(2012 年),已获授权)和 Plasmaviridae(来自 Poddar 等人(1985 年),已获授权)除外。
